The Draft Genome of the Grass Carp Released
China is the world’s largest grass carp culturing country. As one of the most important freshwater aquaculture species in the world, grass carp make up 16 percent of the world’s cultured fish. Most of the freshwater aquaculture species in China belongs to the cyprinidae fish, as well as the grass carp.
Now, three teams led by Prof. Zhu Zuoyan and Prof. Wang Yapping at Institute of Hydrobiology, Prof. Bin Han at National Center of Gene Research, Shanghai Institute of Plant Physiology and Ecology (SIPPE), and Prof. Lin HaoRan at Sun Yat-sen University report the draft genome of the grass carp (Ctenopharyngodon idellus).
Using a whole-genome shotgun sequencing strategy and improved de novo Phusion-meta assemble methods, Wang and his colleagues generate a 0.9-Gb draft genome of a gynogenetic female adult and a 1.07-Gb genome of a wild male adult. Gene annotation identifies 27,263 protein-coding genes in the female genome. Comparative genomics analyses demonstrate that zebrafish and grass carp share a similar genomic evolutionary history. Divergence between grass carp and zebrafish is estimated to have occurred 49–54 million years ago.
More interestingly, Wang and his colleagues reveal that transcriptional activation of the mevalonate pathway and steroid biosynthesis in liver is likely associated with the grass carp’s adaptation from a carnivorous to an herbivorous diet.
“The genome sequence of cyprinidae fish species will provide key technical support to understanding of important developmental characters and genetic improvement of breeding varieties”, says Prof. Han. Meanwhile, it will also benefit the studies of genome evolution, sex determination and differentiation mechanism in fishes.
This work entitled “The draft genome of the grass carp (Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation” was published in Nature Genetics on May 4th, 2015.
This work is supported by the National High-Technology Research and Development Program (863 Program, 2011AA100403), the National Natural Science Foundation of China (31130055), the Strategic Pilot Science and Technology Projects (A) Category, Chinese Academy of Science (XDA08030203), the Guangdong Provincial Science and Technology Program (2012B090500008) and the State Key Laboratory of Freshwater Ecology and Biotechnology (2011FBZ18).
CONTACT:
HAN Bin
National Center of Gene Research, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences
Shanghai 200032, China.
Phone: 86-21-54971325
E-mail:
bhan@ncgr.ac.cn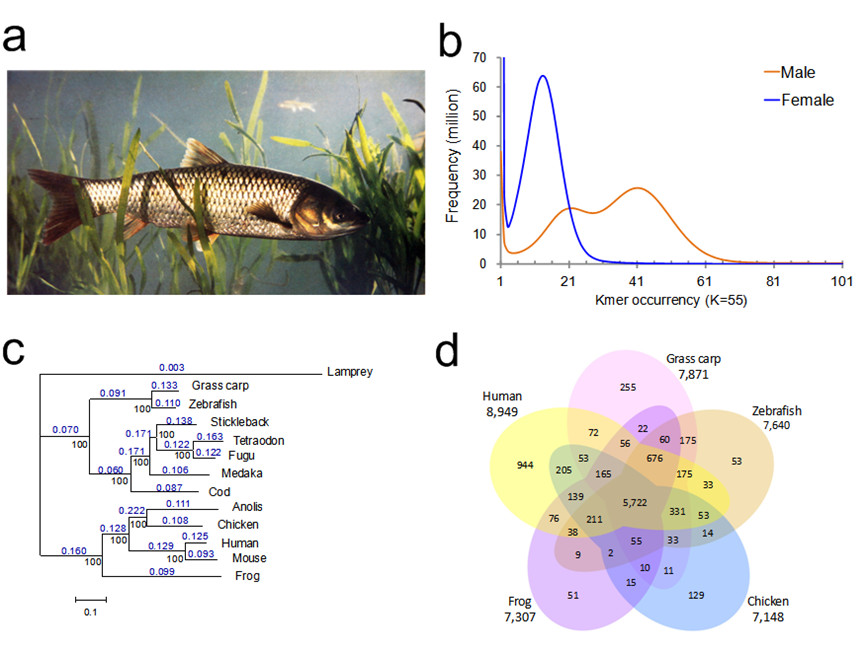
Figure 1 Assemblies and evolution of the grass carp genome. (a) Image of a grass carp adult. (b) Distribution of 55-mer frequency. The distribution of K-mer frequency in the reads was derived from libraries of short insert size (350–400 bp). Values for K-mers are plotted against the frequency (y axis) of their occurrence (x axis). The leftmost truncated peak at low occurrence (1–2) was mainly due to random base errors in the raw sequencing reads. (c) Reconstructed phylogeny of 13 vertebrate genomes. The dN/dS ratio of each branch is shown in blue. The numbers in black correspond to values of bootstrap support. The lamprey is used as an outgroup. Branch length is measured in expected substitutions per site. (d) Venn diagram of gene clusters for five selected vertebrate genomes. Each number represents the number of orthologous gene families shared by the indicated genomes.
