Conserved Intronic Sequences Promote Trans-splicing in Higher Eukaryotes
RNA splicing is one of the essential steps of RNA processing during eukaryotic gene expression and regulation. Unlike the typical cis-splicing, trans-splicing joins exons from two separate transcripts to produce chimeric mRNA and has been detected in most eukaryotes. Trans-splicing in trypanosomes and nematodes has been characterized as a spliced leader RNA-facilitated reaction. However, its mechanism in higher eukaryotes remains unclear.
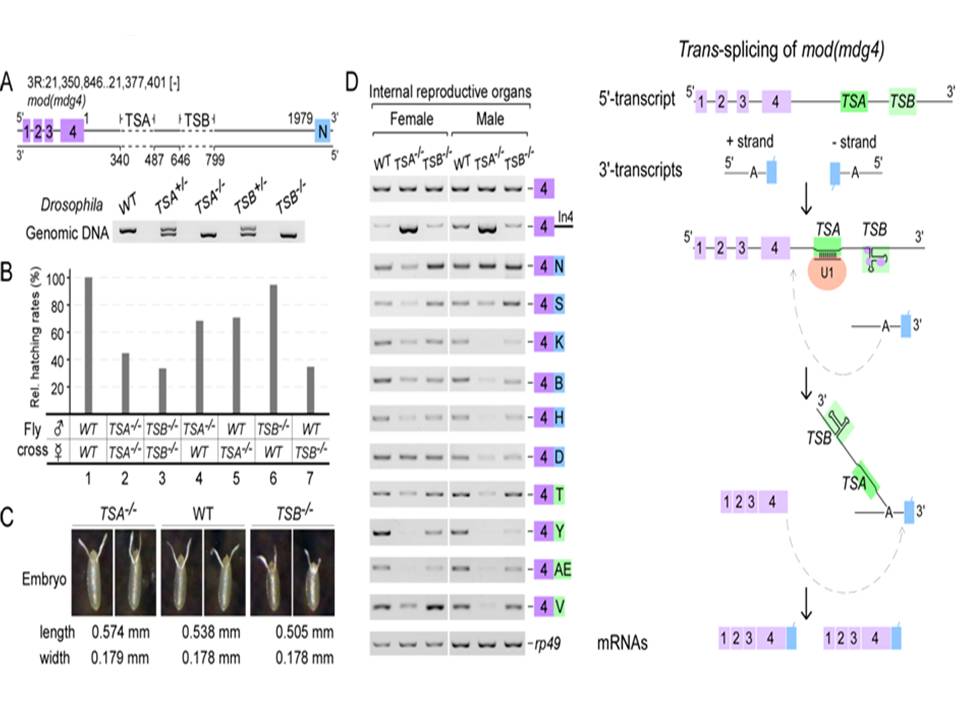
In a recent study, graduate students GAO Jun-Li and WANG Xiu-Ye, and Dr. FAN Yu-Jie under the supervision of Dr. XU Yong-Zhen at the Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, found two critical intronic RNA sequences, TSA and TSB, to promote the trans-splicing of mod (mdg4), a classic trans-spliced gene in Drosophila. TSA, a 13-nt core motif is conserved across Drosophila species and is essential and sufficient for trans-splicing, which binds U1 snRNP through strong base-pairing with U1 snRNA. TSB forms a conserved secondary structure and acts as an enhancer, which binds chromatin factors and U2 snRNP components. Precise deletions of TSA and TSB using the CRISPR/Cas9 system result in developmental and viability defects in flies. Compensatory changes in U1 snRNA rescue trans-splicing of TSA mutants, demonstrating that U1 recruitment is critical to promote trans-splicing in vivo. Furthermore, TSA core-like motifs are also found in many other trans-spliced genes in Drosophila, including lola.
These findings revealed a novel and general mechanism of trans-splicing in higher eukaryotes that the intronic RNA motifs recruit spliceosomal factors and bring separate transcripts into close proximity to promote trans-splicing.
This work entitled “A conserved intronic U1 snRNP-binding sequence promotes trans-splicing in Drosophila” was published in Genes & Development on April 1, 2015.
This research was supported by the grants from the National Natural Science Foundation of China (31270842) and the National Basic Research Program of China (2012CB114101) to XU Y.Z.